Research in our lab aims to understand the biology of microRNAs. These small RNA molecules are important regulators of gene expression in most eukaryotic species. In humans, specific microRNAs are produced in different tissues and at different times during development. Together, they are thought to regulate >60% of mRNAs, and are important in disease. MicroRNAs generally function by binding to partially complementary sites in the 3′ untranslated region (UTR) of target mRNAs, leading to reduced production of the encoded protein.
Regulation of HCV by miR-122
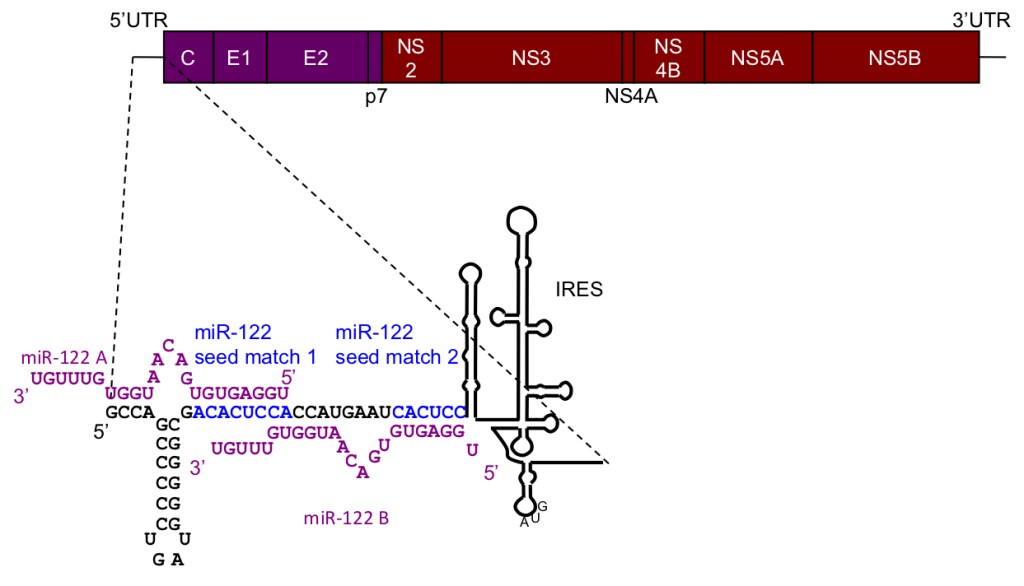
We are particularly interested in miR-122, which is highly and specifically expressed in liver where it constitutes 70% of the total microRNA pool. Previous work by Catherine as a postdoc in Peter Sarnow’s lab at Stanford showed that miR-122 is essential for replication of hepatitis C virus (HCV), which it regulates by an unusual interaction with a tandem binding site in the 5’UTR of the viral RNA (Jopling et al Science 2005).
The mechanism by which miR-122 regulates HCV is still not fully understood. We are working to elucidate this mechanism by identifying the protein factors involved (Ahmed et al NAR 2018, Roberts et al NAR 2014, Roberts et al NAR 2011).
MicroRNA biogenesis from lncRNAs
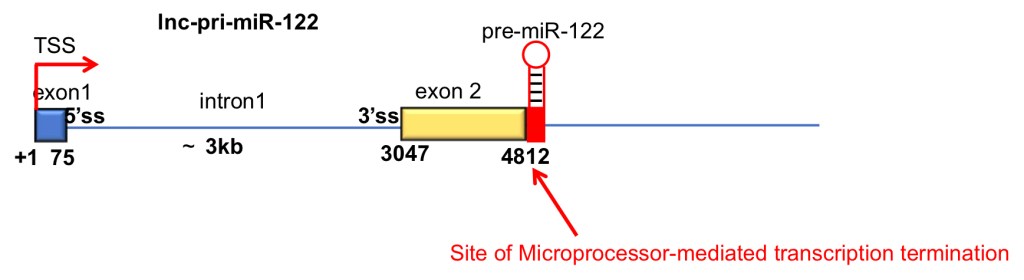
MicroRNAs are expressed as part of longer primary (pri)-miRNA transcripts, which are processed in the nucleus by the Microprocessor complex (Drosha+DGCR8) and then in the cytoplasm by Dicer to generate a mature miRNA. Pri-miR-122 is a long noncoding (lnc)RNA. In collaboration with Nick Proudfoot’s group in Oxford, we found that pri-miR-122 and other lnc-pri-miRNAs use an unusual mechanism of transcription termination which is mediated directly by Microprocessor cleavage, in contrast to the usual mechanism of cleavage and polyadenylation (Dhir et al, NSMB 2015).
We were recently awarded a grant by the BBSRC to investigate the factors that drive this unusual transcription termination mechanism, and to establish how splicing influences microRNA biogenesis from lncRNAs.
Subcellular localisation of microRNA activity
We are investigating how miR-122 regulation of target mRNAs that are translated at the endoplasmic reticulum compares to that of mRNAs localised in the cytoplasm.
MicroRNA regulation in the inflammatory response
In collaboration with Cornelia de Moor’s group in Nottingham, we are investigating how miRNA regulation of target mRNAs is influenced by changes in initial poly(A) tail length that occur during the inflammatory response.